基于VAE的分子生成
本文将介绍分子生成模型的入门介绍——基于VAE的平面分子生成,通过学习分子特征矩阵实现采样生成分子的功能,项目结构如下:
VAE 2D/
├── data/ # 数据集文件
│ └── ZINC.csv
├── result/ # 结果目录
│ ├── model.pth # 模型权重
│ ├── result.csv # 推理生成的结果
│ └── losses.csv # 平均损失记录
├── config.py # 超参数配置文件
├── dataset.py # 加载数据集
└── train.py # 训练代码本文基于该文章进行了代码优化和改造:
参考文献:MGCVAE: Multi-Objective Inverse Design via Molecular Graph Conditional Variational Autoencoder,文章链接在此处
项目概述
模型的架构流程如下图,编码器和解码器的内部实现是多层线性层:
数据嵌入
该模型使用固定大小的特征矩阵来描述分子特征,为了解决每个分子大小和边数不一致的问题,首先要定义最大支持的分子数为
单个分子的特征由三部分描述,首先是
具体的示意图用阿司匹林分子来演示,如下图:
最后我们可以可视化一下阿司匹林分子的全局特征矩阵,如下图:
代码详解
最后对项目的代码进行详细讲解。
配置文件
首先定义常用的模型超参数、相关文件的保存路径。
import torch
import torch.nn as nn
from rdkit import Chem
from rdkit.Chem import rdchem
from pathlib import Path
result_path = Path('./result')
result_path.mkdir(exist_ok=True, parents=True)
# 数据集路径
DATA_FILE = "./data/ZINC.csv"
# 权重保存文件
PT_PATH = "./result/model.pth"
# 损失保存路径
LOSS_PATH = "./result/losses.csv"
# 推理生成结果文件
RESTULT_PATH = "restult.csv"
# 测试集/训练集划分比例
DIVIDE_RATIO = 0.1
# 要生成的分子数
GEN_NUM = 1000
# 批次大小
BATCH_SIZE = 1024
# 训练周期
EPOCH = 200
# 学习率
LR = 0.00005
# 设备
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"定义支持的原子种类和化学键类型,构建索引列表(将用于独热编码的构建和解析)
# 支持的原子类型列表
ATOM_LIST = ( 'C', 'O', 'N', 'H', 'P', 'S', 'F', 'Cl', 'Br', 'I', 'B', 'Si', 'Sn')
ATOM_LIST = [0] + [Chem.Atom(x).GetAtomicNum() for x in ATOM_LIST]
# 支持的原子种类数
ATOM_LEN = len(ATOM_LIST)
# 支持的键类型列表
BOND_LIST = (
rdchem.BondType.ZERO,
rdchem.BondType.SINGLE,
rdchem.BondType.DOUBLE,
rdchem.BondType.TRIPLE,
rdchem.BondType.AROMATIC
)
# 支持的键类型数
BOND_LEN = len(BOND_LIST)
# 索引和实际类型对照表
atom_encoder_m = {l: i for i, l in enumerate(ATOM_LIST)}
atom_decoder_m = {i: l for i, l in enumerate(ATOM_LIST)}
bond_encoder_m = {l: i for i, l in enumerate(BOND_LIST)}
bond_decoder_m = {i: l for i, l in enumerate(BOND_LIST)}定义模型的超参数,这里使用两个线性层来进行特征提取,将输入特征压缩到隐空间维度。
# VAE参数配置
# Encoder: IN_DIM -> H1_DIM -> H2_DIM -> Z_DIM
# Decoder: Z_DIM -> H2_DIM -> H1_DIM -> IN_DIM (out)
# 最大原子数
MAX_SIZE = 16
# 分子特征矩阵列数
F_COL = 1 + ATOM_LEN + MAX_SIZE * BOND_LEN
# 输入维度
IN_DIM = MAX_SIZE * F_COL
# 隐空间维度
Z_DIM = 128
# 隐藏层1
H1_DIM = 512
# 隐藏层2
H2_DIM = 256数据集加载
导入所需要的库文件
import pandas as pd
from rdkit import Chem
import torch
from torch.utils.data import Dataset, DataLoader
from config import *
import numpy as np根据mol对象构建节点特征矩阵,大小
通过先创建全零矩阵,矩阵一行表示一个原子的独热编码;随后遍历该分子的所有原子,根据索引将对应编码位置1,若原子数小于最大原子数,则将剩下的原子设为虚拟原子"0"。
# 生成节点特征矩阵
def node_features(mol, max_length=MAX_SIZE, atom_types=ATOM_LEN):
# 预制全零张量
features = np.zeros([max_length, atom_types], dtype=np.float32)
# 填充真实原子
for i, atom in enumerate(mol.GetAtoms()):
atomic_num = atom.GetAtomicNum()
if atomic_num in atom_encoder_m:
features[i, atom_encoder_m[atomic_num]] = 1.0
# 多出的原子标记为0号(虚拟原子)
actual_atoms = min(mol.GetNumAtoms(), max_length)
features[actual_atoms:, 0] = 1.0
return torch.tensor(features, dtype=torch.float32)根据mol对象生成边特征矩阵,该矩阵大小为
那么,可以先构建三维张量,形状为
随后遍历分子的每一条边,得到边两端的原子索引,单独构建上三角矩阵,根据边类型构建独热编码向量。
最后把原三维张量
# 生成边特征矩阵
def bond_features(mol, bond_encoder_m, max_length=MAX_SIZE):
bond_types = len(bond_encoder_m)
# 预先构建三维张量,所有键均为“非键”编码的独热向量
A = torch.zeros(bond_types, dtype=torch.int32)
A[0] = 1
A = A.expand(max_length, max_length, bond_types).clone()
# 遍历每个边
for bond in mol.GetBonds():
begin = bond.GetBeginAtomIdx() # 键的起始原子索引
end = bond.GetEndAtomIdx() # 键的终止原子索引
i = min(begin, end) # 行号=原子对中较小的索引
j = max(begin, end) # 列号=原子对中较大的索引
# 构建边类型独热编码向量,替换原值
bond_code = bond_encoder_m[bond.GetBondType()]
bond_onehot = torch.zeros(bond_types, dtype=torch.int32)
bond_onehot[bond_code] = 1
A[i, j] = bond_onehot
# 将三维矩阵按最后一维展开,得到二维特征矩阵
A = A.reshape(max_length, -1)
return A构建分子的全局特征,将上述节点特征、边特征和分子的原子数独热编码向量拼接,得到总特征矩阵,形状为
# mol对象生成分子特征矩阵
def mol_to_graph(mol, length, smiles='', max_num=MAX_SIZE):
try:
mol_length = int(length) - 1
vec_length = torch.zeros([max_num,1], dtype=torch.float)
vec_length[mol_length, 0] = 1.0
graph = torch.cat([vec_length, node_features(mol, max_num),bond_features(mol, bond_encoder_m)], 1).float()
return graph
except Exception as e:
print('Error:', smiles, 'more information:', e)读取csv文件,将smiles转为mol对象,再利用上述mol_to_graph函数将mol对象构建为特征矩阵。
# 读取数据集源文件
df = pd.read_csv(DATA_FILE)
# 按设定大小抽样
df = df[df['Length'] <= MAX_SIZE].reset_index(drop=True)
# 生成mol对象列
df["mol"] = df["SMILES"].apply(lambda x: Chem.MolFromSmiles(x))
# 去除空值
df = df[df["mol"].notna()].reset_index(drop=True)
# 生成特征矩阵列
df['graph'] = df[['mol', 'Length', 'SMILES']].apply(
lambda row: mol_to_graph(mol=row['mol'], length=row['Length'], smiles=row['SMILES']),
axis=1
)
# 去除空值
df = df[df['graph'].notna()].reset_index(drop=True)
# 单独保存特征矩阵列为列表
data_list = df['graph'].to_list()最后定义数据集类,划分数据集,构建数据集加载器(和之前相同)
# 定义数据集类
class MolGraphDataset(Dataset):
def __init__(self, data_list):
self.data_list = data_list
def __len__(self):
return len(self.data_list)
def __getitem__(self, idx):
return self.data_list[idx]
# 初始化自定义数据集
mol_dataset = MolGraphDataset(data_list)
# 划分训练/测试集
tr = 1 - DIVIDE_RATIO
train_size = int(len(mol_dataset) * tr)
test_size = len(mol_dataset) - train_size
# 随机划分
train_dataset, test_dataset = torch.utils.data.random_split(
mol_dataset,
[train_size, test_size],
generator=torch.Generator().manual_seed(42)
)
# 构建DataLoader
train_loader = DataLoader(
dataset=train_dataset,
batch_size=BATCH_SIZE,
shuffle=True,
drop_last=True,
)
test_loader = DataLoader(
dataset=test_dataset,
batch_size=BATCH_SIZE,
shuffle=False,
drop_last=True,
)进行单元测试验证:
if __name__ == "__main__":
print(f"Train dataset size: {len(train_dataset)} samples")
print(f"Test dataset size: {len(test_dataset)} samples")
print(f"Train loader batch count: {len(train_loader)}")
print(f"Test loader batch count: {len(test_loader)}")
# 测试加载单个批次
for batch in train_loader:
print(f"Batch shape: {batch.shape}")
break得到如下结果,获取构建的数据集形状和批次形状。
Train dataset size: 90000 samples
Test dataset size: 10000 samples
Train loader batch count: 87
Test loader batch count: 9
Batch shape: torch.Size([1024, 16, 95])训练与推理
先导入相关的库,禁用rdkit警告
import numpy as np
import pandas as pd
from rdkit import Chem
from rdkit.Chem.Crippen import MolLogP, MolMR
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from config import *
# 禁用rdkit警告
from rdkit import RDLogger
RDLogger.DisableLog('rdApp.*')
import warnings
warnings.filterwarnings(action='ignore')根据节点特征矩阵+边特征矩阵构建新的mol对象,完成合理性检验后返回。
# 分子特征矩阵转为mol对象
def graph_to_mol(node_labels, adjacency, atom_decoder_m, bond_decoder_m, strict=False):
mol = Chem.RWMol()
for node_label in node_labels:
mol.AddAtom(Chem.Atom(atom_decoder_m[node_label]))
for start, end in zip(*np.nonzero(adjacency)):
if start < end:
mol.AddBond(int(start), int(end), bond_decoder_m[adjacency[start, end]])
if strict:
try:
Chem.SanitizeMol(mol)
except:
mol = None
return mol定义推理函数,根据设定的生成分子总数一次生成多个分子矩阵,按输入规则将每个分子特征矩阵拆分为原子数编码+节点特征矩阵+边特征矩阵,再用同样的规则解析即可。
# 解析推理得到的数据
def vae_results(
vae,
atom_decoder_m,
bond_decoder_m,
gen_num=GEN_NUM,
z_dim=Z_DIM,
atom_types=ATOM_LEN,
bond_types=BOND_LEN,
row_dim=MAX_SIZE,
col_dim=F_COL,
):
with torch.no_grad():
z = torch.randn(int(gen_num), z_dim).to(DEVICE)
sample = vae.decoder(z)
smi = []
logp = []
mr = []
for matrix in sample.view(int(gen_num), 1, row_dim, col_dim).cpu():
try:
# 单个分子特征矩阵
matrix = matrix[0]
# 原子数编码矩阵
atom_num = matrix[:, 0]
# 独热转特征
atom_num = torch.argmax(atom_num).item() + 1
# 原子类型编码矩阵
atom_type = matrix[:,1:atom_types+1]
# 原子类型独热编码转类型索引
atom_type = torch.max(atom_type, -1)[1]
# 边特征编码矩阵
bonds = matrix[:, atom_types+1:]
# 边特征独热编码转特征索引的邻接矩阵
bonds = bonds.reshape(row_dim, row_dim, bond_types)
bonds = torch.max(bonds, dim=-1)[1]
# 只保留有效原子部分
nodes_hard_max = atom_type[:atom_num]
edges_hard_max = bonds[:atom_num, :atom_num]
# 从特征矩阵解码还原分子mol对象
mol = graph_to_mol(
nodes_hard_max.numpy(),
edges_hard_max.numpy(),
atom_decoder_m,
bond_decoder_m,
strict=True
)
# 有效性检查
if mol and '.' not in Chem.MolToSmiles(mol):
smi.append(Chem.MolToSmiles(mol))
logp.append(MolLogP(mol))
mr.append(MolMR(mol))
except Exception as e:
print(e)
continue
return pd.DataFrame({'SMILES': smi, 'logP': logp, 'MR': mr})根据模型输出矩阵和原始矩阵,计算BCE损失和KL散度损失,作为总损失返回。
# 损失函数
def loss_function(recon_x, x, mu, log_var):
# BCE损失
BCE = F.binary_cross_entropy(recon_x, x.view(-1, IN_DIM), reduction='sum')
# KL散度损失
KLD = -0.5 * torch.sum(1 + log_var - mu.pow(2) - log_var.exp())
return BCE + KLD模型的架构与之前的图像VAE基本一致:
# 模型定义
class VAE(nn.Module):
def __init__(self, x_dim=IN_DIM, h_dim1=H1_DIM, h_dim2=H2_DIM, z_dim=Z_DIM):
super(VAE, self).__init__()
self.x_dim = x_dim
# 编码器
self.fc1 = nn.Linear(x_dim, h_dim1)
self.fc2 = nn.Linear(h_dim1, h_dim2)
self.fc31 = nn.Linear(h_dim2, z_dim)
self.fc32 = nn.Linear(h_dim2, z_dim)
# 解码器
self.fc4 = nn.Linear(z_dim, h_dim2)
self.fc5 = nn.Linear(h_dim2, h_dim1)
self.fc6 = nn.Linear(h_dim1, x_dim)
def encoder(self, x):
h = F.relu(self.fc1(x))
h = F.relu(self.fc2(h))
return self.fc31(h), self.fc32(h)
def sampling(self, mu, log_var):
std = torch.exp(0.5*log_var)
eps = torch.randn_like(std)
return eps.mul(std).add(mu)
def decoder(self, z):
h = F.relu(self.fc4(z))
h = F.relu(self.fc5(h))
return torch.sigmoid(self.fc6(h))
def forward(self, x):
mu, log_var = self.encoder(x.view(-1, self.x_dim))
z = self.sampling(mu, log_var)
return self.decoder(z), mu, log_var实现训练+测试函数
# 训练函数
def train(model, train_loader):
train_loss = 0
model.train()
for graph in train_loader:
graph = graph.to(DEVICE)
optimizer.zero_grad()
recon_batch, mu, log_var = model(graph)
loss = loss_function(recon_batch, graph, mu, log_var)
loss.backward()
optimizer.step()
train_loss += loss.item()
return train_loss
# 测试函数
def test(model, test_loader):
test_loss= 0
model.eval()
with torch.no_grad():
for graph in test_loader:
graph = graph.to(DEVICE)
recon, mu, log_var = model(graph)
test_loss += loss_function(recon, graph, mu, log_var).item()
return test_loss最后初始化模型,开始训练,完成后保存权重,进行推理即可。
if __name__ == "__main__":
# 定义模型,迁移设备
vae = VAE()
vae.to(DEVICE)
# 设置优化器
optimizer = optim.Adam(vae.parameters(), lr=LR)
print(f'Train device:{DEVICE}')
print('Load Dataset......')
# 载入数据集
from dataset import train_loader, test_loader
print('='*50)
print(f"Train dataset size: {len(train_loader.dataset)} samples")
print(f"Test dataset size: {len(test_loader.dataset)} samples")
print(f"Train loader batch count: {len(train_loader)}")
print(f"Test loader batch count: {len(test_loader)}")
print('='*50)
print('Training the model...')
# 开始迭代训练周期
train_loss_list = []
test_loss_list = []
for epoch in range(1, EPOCH+1):
# 训练周期损失
train_loss = train(vae, train_loader) / len(train_loader.dataset)
train_loss_list.append(train_loss)
# 测试周期损失
test_loss = test(vae, test_loader) / len(test_loader.dataset)
test_loss_list.append(test_loss)
print(f'Epoch [{epoch}/{EPOCH}], train loss {train_loss:.4f}; val loss {test_loss:.4f}')
# 保存权重
torch.save(
vae.state_dict(),
PT_PATH
)
print(f"Saving PTH to:{PT_PATH}")
# 保存每轮损失
import pandas as pd
df = pd.DataFrame({
'Epoch': range(1, len(train_loss_list) + 1),
'Training Loss': train_loss_list,
'Test Loss': test_loss_list
})
df.to_csv(LOSS_PATH, index=False)
print(f"Saving Loss to:{LOSS_PATH}")
# 加载权重
vae.load_state_dict(torch.load(PT_PATH, map_location=DEVICE), strict=True)
print('Generating molecules...')
vae_df = vae_results(vae, atom_decoder_m, bond_decoder_m)
vae_df.to_csv(RESTULT_PATH, index=False)
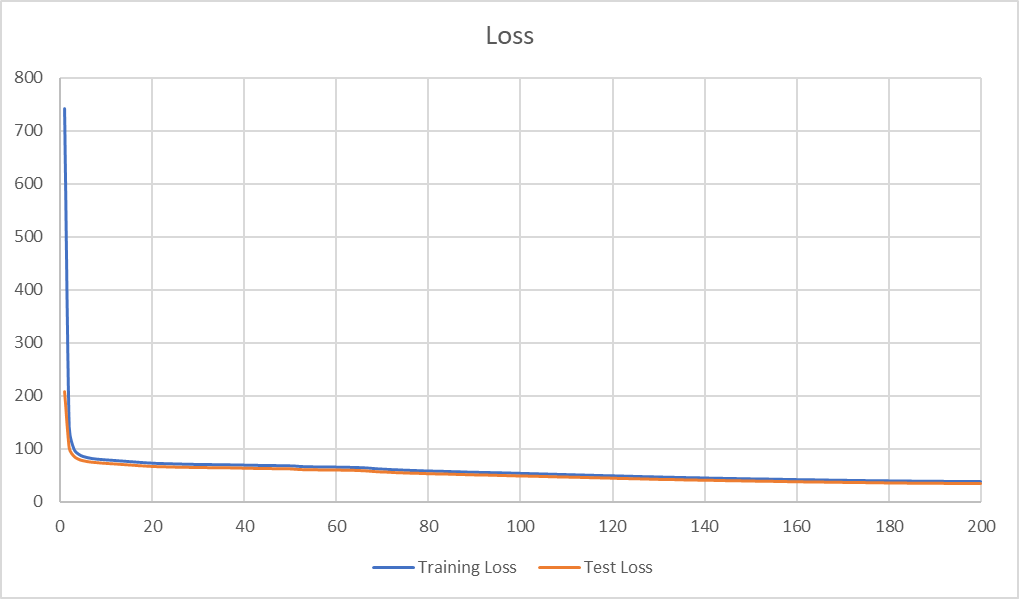
print(f'Saving {RESTULT_PATH} ({vae_df.shape[0]})...')最终的训练结果如下:
Train device:cuda
Load Dataset......
==================================================
Train dataset size: 90000 samples
Test dataset size: 10000 samples
Train loader batch count: 87
Test loader batch count: 9
==================================================
Training the model...
Epoch [1/200], train loss 747.0516; val loss 208.7150
Epoch [2/200], train loss 141.5465; val loss 101.5751
Epoch [3/200], train loss 99.4068; val loss 86.2277
......
Epoch [196/200], train loss 37.5508; val loss 35.0344
Epoch [197/200], train loss 37.5112; val loss 35.0496
Epoch [198/200], train loss 37.4262; val loss 34.9374
Epoch [199/200], train loss 37.3526; val loss 34.8599
Epoch [200/200], train loss 37.3289; val loss 34.8170
Saving PTH to:./result/model.pth
Saving Loss to:./result/losses.csv
Generating molecules...
Saving ./result/restult.csv (323)...观察损失曲线:
模型推理得到的分子列表如下:
SMILES,logP,MR
CC1(C(=O)NCCCN)CO1,-0.7596999999999994,41.03810000000001
CC(=O)NC(=O)NC(=O)CCCO,-0.8687999999999998,43.67520000000001
COC1CC1(C)NNCC(C)O,-0.36119999999999997,46.63820000000002
CCCNCC(C)NCN(C)C,0.4831999999999999,54.31240000000004
CCOC(O)C(C)C(C)CO,0.6057999999999999,43.04660000000001
CCC(CCO)C(C)CCCO,1.8035999999999999,50.96760000000003
CC(=O)CCCCC(C)CC(O)O,1.4726999999999997,51.155600000000035
CC(CO)NC(=O)CCCNC=O,-0.9903999999999991,47.99120000000003本文中我们基于VAE架构实现了最简单的二维分子生成模型,该模型还有许多需要改进的地方,后续我们将对其进行改造,实现条件生成的功能。
其他的,还可以引入图数据结构,用GNN做特征提取,结合图解码器实现生成。后续模型详见本站其他文章。